 PhD (FPI) Contract
PhD (FPI) Contract
Contrato FPI (Formación de Personal Investigador) para realizar la tesis doctoral.
PhD contract (4-year FPI contract)
Research Project: Adaptive evolution in insular radiations of spiders from genus Dysdera (Arachnida: Dysderidae) using data from high quality complete genomes.
PID2022-138477NB-C22
IP: Julio Rozas (Universitat de Barcelona; Barcelona).
This project aims to study the genome evolution of the spider genus Dysdera and its diversification in the Canary Islands.
We will investigate the genomic basis of the adaptation of several species of this group to feed preferentially (facultatively or even obligatory) woodlice,
a clear example of convergent phenotypic evolution associated with a specialist diet).
We will also use this case example to gain insight into the molecular evolution of some important gene families in chelicerates,
such as those encoding the molecular components of the chemosensory system and the venom in these spiders.
The student will participate in the assembly, identification, annotation and evolutionary (comparative and population) genomic analysis using data from complete (including chromosome-level) genomes.
For that, he/she will use high quality genome sequences, bioinformatics tools (software and scripts to manipulate and visualizate sequences and genomic annotations, to identify gene family copies).
The data will be analyse using phylogenetic and evolutionary genetic methods, under the theoretical framework of population genetics and molecular evolution.
Many of these analyses will be carried out in our high performance computer cluster.
The project will generate high-quality genomic data based on third-generation sequencing platforms,
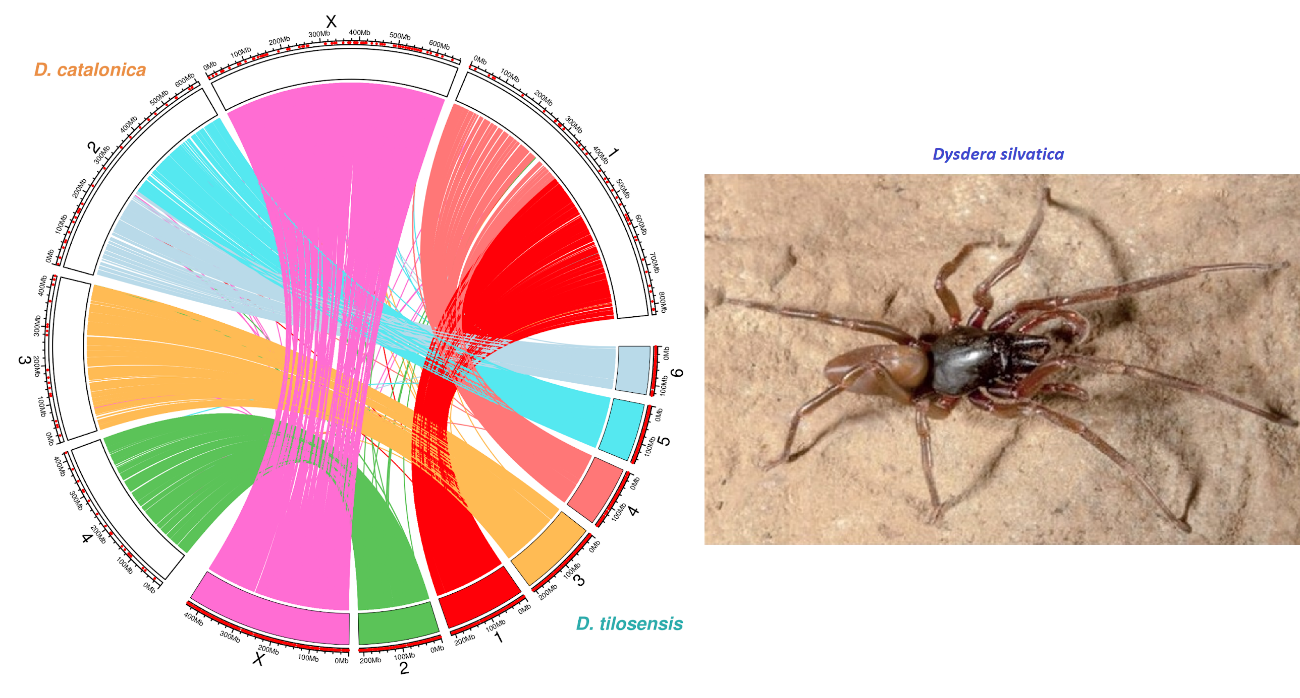
which will be the source to carry out comparative and population genomic studies in six endemic Dysdera species from the Canary Islands, as well as in two continental species.
The chromosomal-level high-quality assemblies will allow us characterizing the adaptive genomic architecture of this extraordinary radiation while provide new and valuable knowledge on the evolutionary genomics of ecological adaptations and the repetitiveness of evolution.
Furthermore, these new data are a great opportunity 1) to accurately characterize relevant gene families (such as those encoding the chemosensory system) and repetitive elements for the first time in high-quality chelicerate genomes, and
2) to study the impact of the divergent vs convergent evolution (including the analysis of gene conversion) on the gene family evolution.
For the analysis, we will develop, implement, and apply bioinformatics tools and specific software for the manipulation and analysis of massive sequencing data;
the new developed tools will be especially useful for researches working on non-model organisms.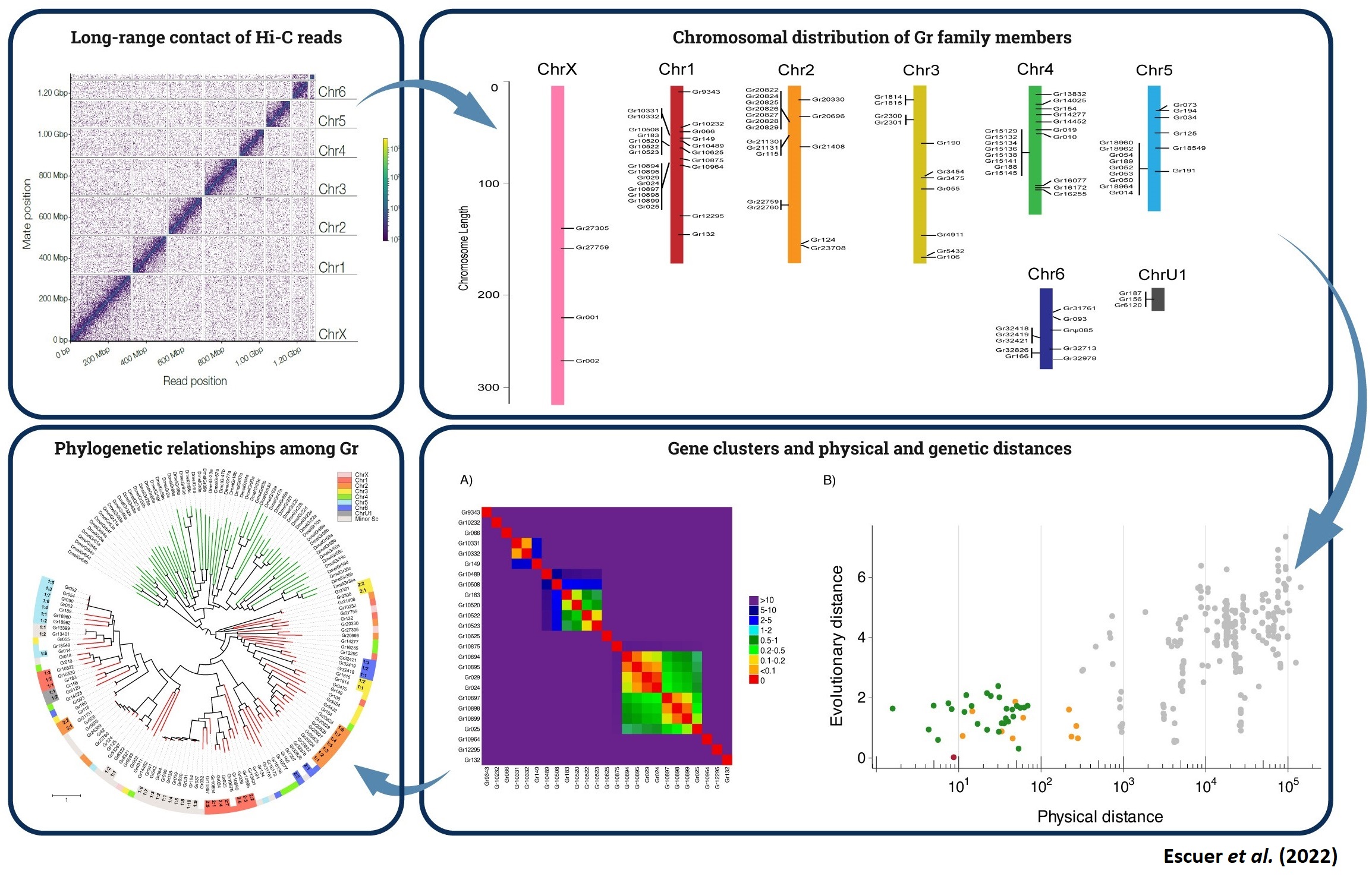
Coordinated Project:
This project is coordinated with PID2022-138477NB-C21 (PI: Rafael Zardoya; MNCN, Museo Nacional de Ciencias Naturales-CSIC), which using similar genomic and bioinformatic tools aims at deciphering the origin and evolution of multigene families in venomous marine cone snails, with particular emphasis on conotoxin and chemosensory gene families. Given the common framework of both coordinated projects, it is expected that fruitful collaborations will be established (including short stays in MNCN), in which the student will actively participate.Expected skills:
Knowledge about comparative genomics, phylogenetics and bioinformatic tools, and/or on NGS data handling, assembly and analysis.Experience with Linux operating systems, and some programing languages commonly used in bioinformatics (Perl, Python, R).
Requeriments and Deadlines:
Candidates must have a master degree in Life Sciences.Please send ASAP (by september 15th, 2023) a single email to Julio Rozas, including:
1) a brief CV, 2) your BS and MS transcripts with your academic grades, and 3) a motivation letter.
Indicate in the email subject: FPI2023 -Your name
The selection process will start on the second-third week of september-2023. Incorporation date. About January 2024.
References and Publications:
Julio Rozas' Publications
Contact: jrozas@ub.edu
Looking for a postdoc? Interested in Comparative/Population Genomics at our lab? You may be interested in applying to next JdC (Juan de la Cierva), or other upcoming calls.
Software developed in the research group:
Publications of the EGB research group:
PostDoc Opportunities