Welcome CBDD
WELCOME to Computational Biology and Drug Design (CBDD) group
The CBDD is a research group pertaining to the Faculty of Pharmacy and Food Sciences, and their members are ascribed to the research centers:
- the Institute of Theoretical and Computational Chemistry (IQTCUB),
- the Institute of Biomedicine (IBUB), and
- the Institute for Nutrition and Food Safety Research (INSA).
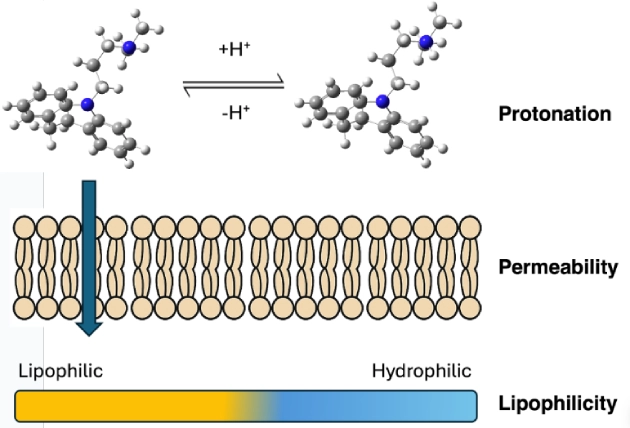
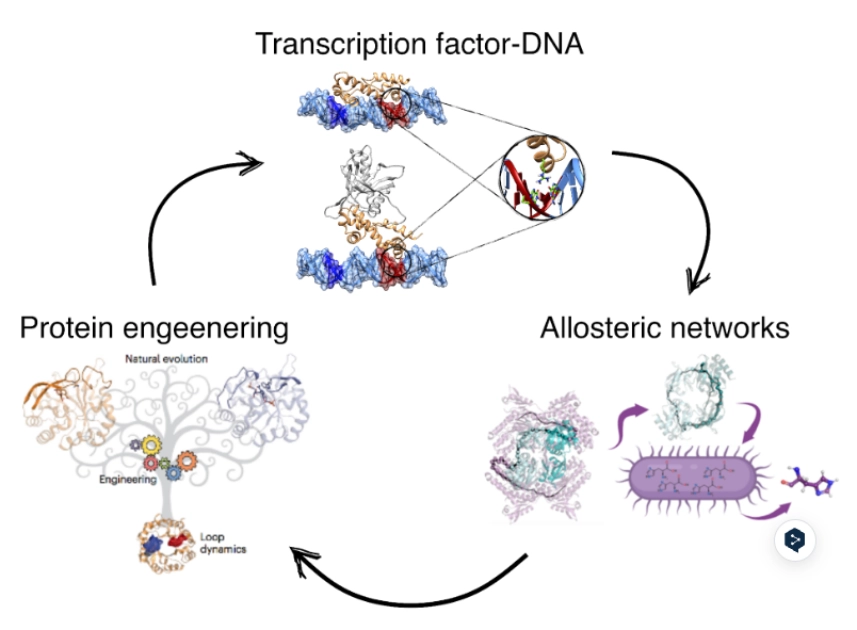
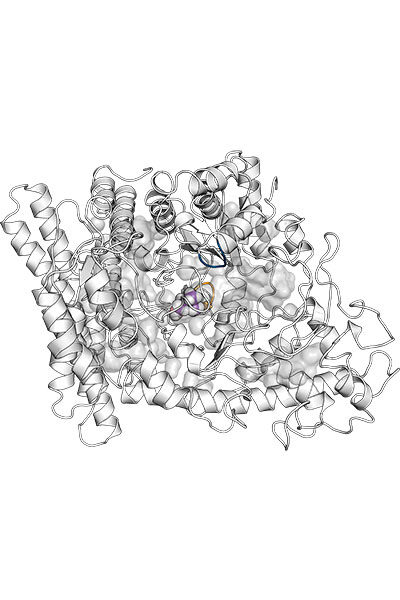
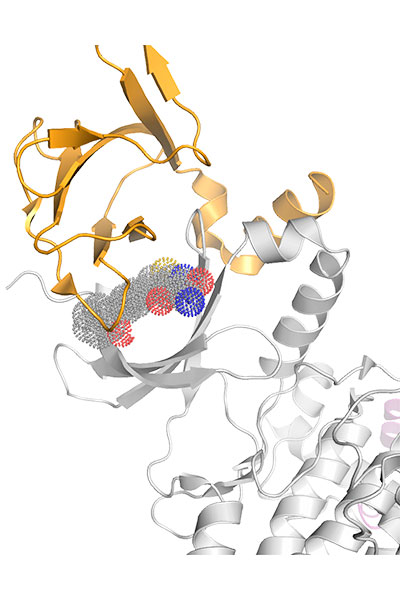
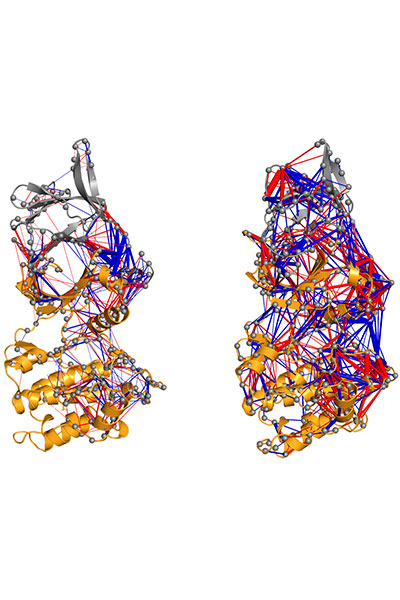
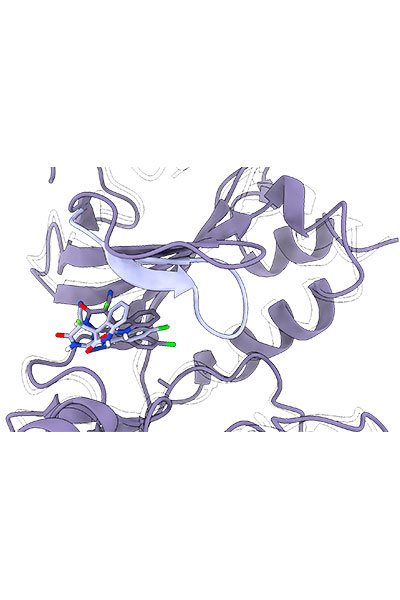
Our research is focused on the study of the chemical properties of small (bio)organic molecules and their recognition and binding with macromolecular targets. Emphasis is made in understanding the molecular determinants of biomolecular association, the relationships between structure and biological function, and the design of novel bioactive compounds, especially regarding the application in drug discovery.
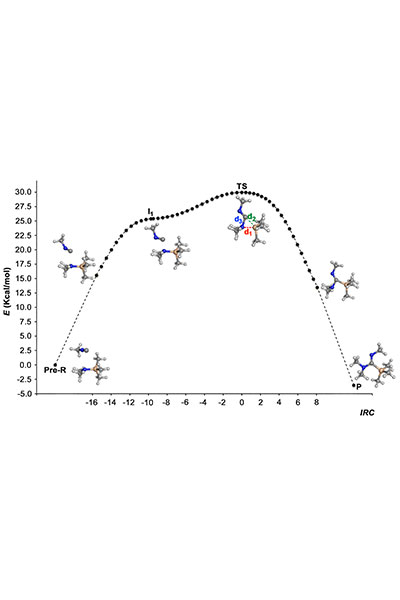
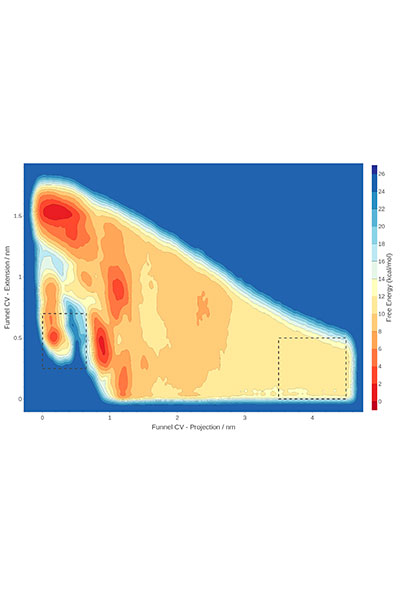
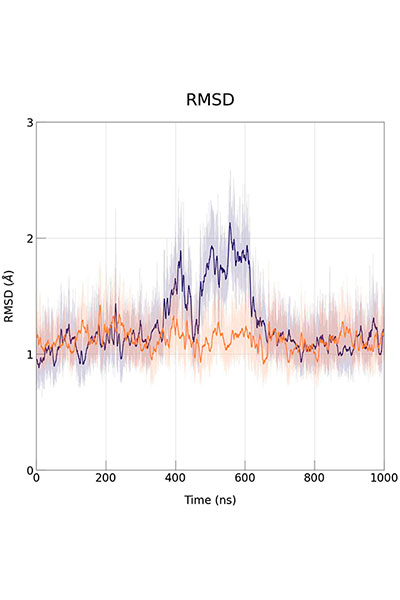
Our research is mainly performed by using a variety of tools afforded by theoretical and computational methods of molecular modeling, quantum chemistry, classical simulations, advanced sampling techniques, and computer-aided drug design. Nevertheless, we have consolidated over the years a crosstalk with experimental groups in chemical synthesis and biophysical studies.
Furthermore, our research also comprises the development of novel methodologies for the study of molecular recognition and drug design, which is mainly done in collaboration with Pharmacelera (https://pharmacelera.com/).