Comparative genomics of gene expression lab
Department of Genetics, Microbiology and Statistics
Faculty of Biology, UB
Marco Mariotti
Ramon y Cajal Researcher
ORCID ID: 0000-0002-7411-4161
https://www.mariottigenomicslab.com/
Principal investigator: • Marco Mariotti
PhD students: • Nadezhda (Nadya) Makarova
Technician: • Sergio Sanchez Moragues

Current
Research
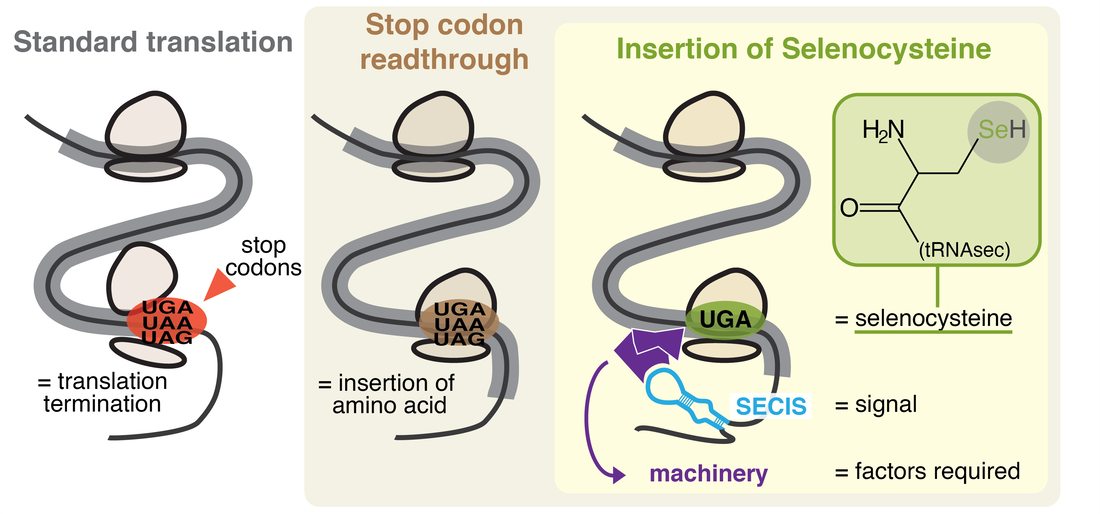
Certain genes are translated with programmed exceptions to the genetic code used for the rest of the genome. This phenomenon is known as “translational recoding”, and it is widespread through life. Its most common form is stop codon readthrough, wherein a stop codon supports amino acid insertion instead of translation termination.
Selenocysteine (Sec) is a non-canonical amino acid found in oxidoreductases known as selenoproteins. Sec is encoded by the UGA codon, and its insertion is dependent on stop codon readthrough. Humans carry 25 selenoproteins, many of which have essential functions such as antioxidant defense, protein quality control, and hormone regulation. This makes Sec, and its component selenium, essential for human health.
Our lab investigates the biology of selenium utilization and selenoproteins. We are particularly interested in two selenoproteins central to selenium metabolism: selenophosphate synthetase (Sephs2), involved in Sec synthesis, and Selenoprotein P (Sepp1), involved in selenium transport.
In insects, selenoproteins are very rare, but other forms of stop codon readthrough are very abundant: evolutionary conservation analyses indicate that readthrough occurs in ~1000 Drosophila genes. The functions and mechanisms of this phenomenon, however, are poorly characterized.
Our lab investigates stop codon readthrough in insects through computational and experimental approaches. We detect occurrences of readthrough through analysis of ribosome profiling, which is a variant of high-throughput RNA sequencing. Then, we pinpoint and validate the sequence elements responsible for readthrough through reporter assays in cell culture.
Besides our main basic research goals, we are working on repurposing natural readthrough mechanisms for an innovative form of gene therapy. Our aim is to induce readthrough in a targeted manner, which will allow to rescue pathogenic non-sense mutations.
Selected
Publications
Manta, B.; Makarova, N.E.; Mariotti, M. The selenophosphate synthetase family: a review. Free Radical Biology and Medicine. 16:S0891-5849(22)00589-5, 2022. doi.org/10.1016/j.freeradbiomed.2022.09.007
Santesmasses, D.; Mariotti, M.; Gladyshev, V.N. Tolerance to selenoprotein loss differs between human and mouse. Molecular Biology and Evolution. 37 (2), 341-354, 2020. doi.org/10.1093/molbev/msz218
Mariotti, M.; Salinas, G.; Gabaldón, T.; Gladyshev, V.N. Utilization of selenocysteine in early-branching fungal phyla. Nature Microbiology. 4:759-765, 2019. doi.org/10.1038/s41564-018-0354-9
Mariotti, M.; Shetty, S.; Baird, L.; Wu, S.; Loughran, G.; Copeland, P.R.; Atkins, J.F.; Howard, M.T. Multiple RNA structures affect translation initiation and UGA redefinition efficiency during synthesis of selenoprotein P. Nucleic Acids Research. 45:13004-13015, 2017. doi.org/10.1093/nar/gkx982
Mariotti, M.; Santesmasses, D.; Capella-Gutierrez, S.; Mateo, A.; Arnan, C.; Johnson, R.; D’Aniello, S.; Hee-Yim, S.; Gladyshev, V.N.; Serras, F.; Corominas, M.; Gabaldón, T.; Guigó, R. Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization. Genome Research. 25(9):1256-67, 2015. doi.org/10.1101/gr.190538.115


